I am an evolutionary and molecular biologist with a particular interest on studying host/symbiont interactions, from the phenotypic level to the genomic aspects of the interaction. I have completed my PhD degree in France (Université de Poitiers, December 2012) focusing my research on the immediate and evolutionary consequences of symbiont horizontal transfers on host life-history traits, using the Wolbachia-woodlouse model. I have investigated patterns of virulence to the invertebrate hosts following injection of different strains of the intracellular bacteria Wolbachia from related host species, as well as the impact of transmission mode (vertical vs. horizontal) on the evolution of Wolbachia‘s virulence.
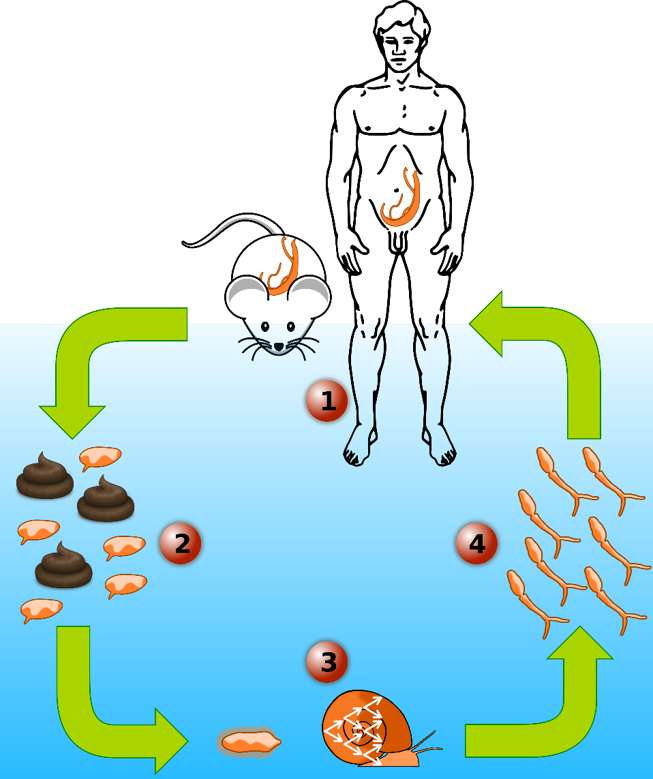
Since March 2013, I am part of Dr. Tim Anderson’s Lab at the Texas Biomedical Research Institute as a Postdoctoral scientist first then as Staff Scientist. My current research focuses on studying the human blood fluke schistosome, the most important of the helminth parasites infecting humans. This parasite infects 200 million people and leading to an estimated 200,000 deaths each year. I aim to decipher the genetic basis of multiple biomedically important traits in this parasite like drug resistance or transmission/virulence traits:
- The genetic basis of virulence/transmission in the human blood fluke, Schistosoma mansoni: using classical genetic crosses approach followed by Next-generation exome sequencing and quantitative genetic, we have identified several regions of the S. mansoni genome involved in this particular trait.
- The genetic basis of praziquantel (PZQ) resistance (the only drug available on the market to treat schistosomiasis) in S. mansoni: using a lab population, polymorphic for PZQ resistance, a genetic association approach followed by bulk segregant analysis, we have pinpointed a QTL involved in PZQ resistance and we are working on functional validation of genes identified.
- A large field population genomic study of S. mansoni and S. haematobium using larval samples directly collected from patients feces/urine in Africa (in collaboration with the SCAN) and South America, to understand the populations structure of field parasites and go on the tracks of the South American colonisation of S. mansoni using exome-wide markers.
- The exome sequencing of several genetic crosses of S. mansoni to refine a genome: using our genetic crosses (F0 to F2), genetic map and linkage analysis we helped to improve and correct the assembly of the S. mansoni reference genome (in collaboration with the Sanger institute).
- The development of many new methods and protocols to improve the molecular toolkits available for Schistosome parasites.
Schistosome snail host microbiome. I am the co-lead of a pioneering schistosome snail host microbiome project, where we investigate the diversity and abundance of microorganisms within the hemolymph (i.e. blood) of Biomphalaria snails. We highlighted significant differences in microbiome composition at the level of individual snails, snail populations and species. We have also established a snail organs microbiome atlas and we are currently investigating the impact of schistosome infections on the snail microbiome. Understanding the role of microbiome in the snail-parasite interaction can lead to a better control of parasite populations, especially if some specific bacteria help snails to resist to the infection.